Review: Nvidia’s RAPIDS brings Python analytics to the GPU
An end-to-end data science ecosystem, open source RAPIDS gives you Python dataframes, graphs, and machine learning on Nvidia GPU hardware
-
RAPIDS v0.11
Building machine learning models is a repetitive process. Often rote and routine, this is a game of “fastest through the cycle wins,” as the faster you can iterate, the easier it is to explore new theories and get good answers. This is one of the reasons practical enterprise use of AI today is dominated by the largest enterprises, which can throw enormous resources at the problem.
RAPIDS is an umbrella for several open source projects, incubated by Nvidia, that puts the entire processing pipeline on the GPU, eliminating the I/O bound data transfers, while also substantially increasing the speed of each of the individual steps. It also provides a common format for the data, easing the burden of exchanging data between disparate systems. At the user level, RAPIDS mimics the Python API in order to ease the transition for that user base.
 The Tidyverse Cookbook
The Tidyverse Cookbook
Typical machine learning workflow
RAPIDS ecosystem architecture
The RAPIDS project aims to replicate, for the most part, the machine learning and data analytics APIs of Python, but for GPUs rather than CPUs. This means that Python developers already have everything they need to run on the GPU, without having to learn the low-level details of CUDA programming and parallel operations. Pythonistas can develop code on a non-GPU enabled machine, then, with a few tweaks, run it on all the GPUs available to them.
The Nvidia CUDA toolkit provides lower level primitives for math libraries, parallel algorithms, and graph analytics. At the heart of the architecture is the GPU data frame, based on Apache Arrow, that provides a columnar, in-memory data structure that is programming language agnostic. The user interacts with the GPU dataframe via cuDF and a Pandas-like API. Dask, a Python library for parallel computing, mimics the upstream Python APIs and works with CUDA libraries for parallel computation. Think of Dask as Spark for Python.
 RAPIDS
RAPIDS
RAPIDS ecosystem architecture
The three main projects, cuDF, cuML and cuGraph, are developed independently, but designed to work seamlessly together. Bridges to the broader Python ecosystem are also being developed as part of the project.
RAPIDS installation
Installation via Anaconda on a Linux machine in AWS was mostly straightforward, barring a few hiccups due to a change in dependencies in version 0.11. Installing the C/C++ libraries to use libcudf was not so easy, and I’d recommend sticking to the Python APIs and Conda installation process. RAPIDS includes a Jupyter notebook, also available on Google’s free Colab, that makes getting started simple. I used the Jupyter notebook version 0.10 to run the code on Google Colab, which includes an Nvidia Tesla T4 GPU.
 IDG
IDGRAPIDS’ GPU dataframe
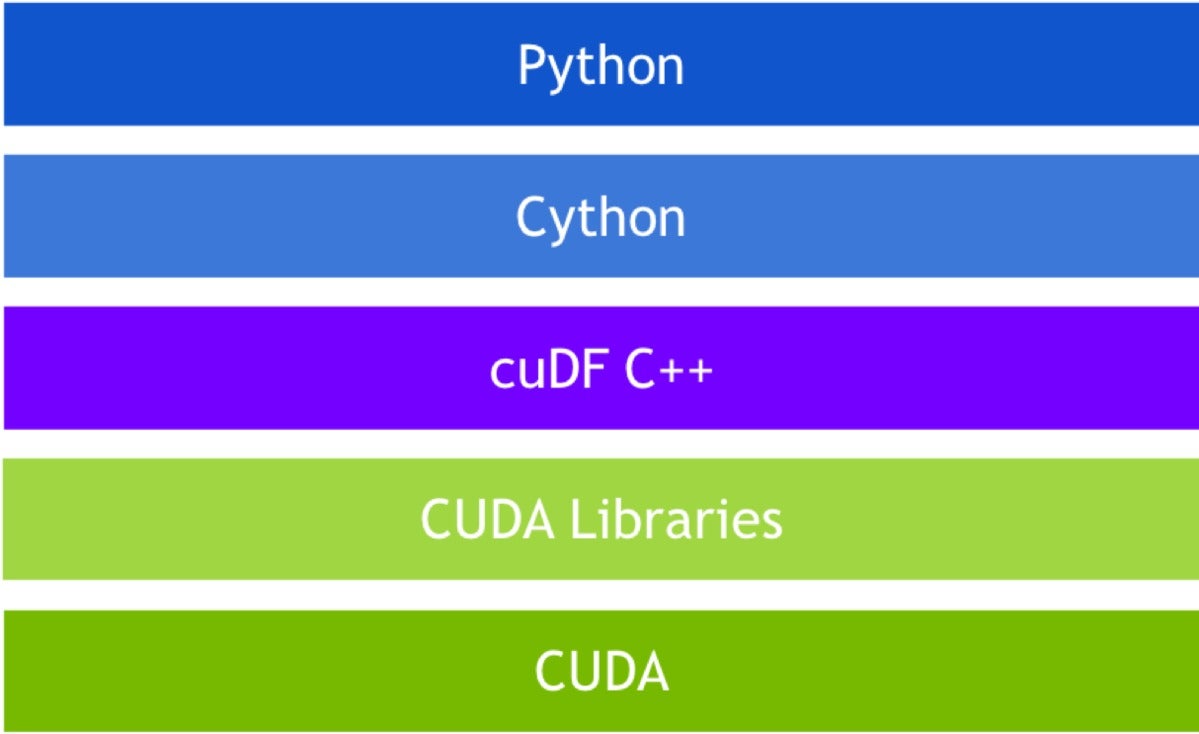
At the heart of any data science workflow is the dataframe. This is where feature engineering happens, and where the majority of time is spent, as data scientists wrangle dirty data. cuDF is the RAPIDS project for a GPU-based, Pandas-like dataframe. Underpinning cuDF is libcudf, a C++ library implementing low-level primitives for importing Apache Arrow data, performing element-wise mathematics on arrays, and executing sort, join, group by, reduction, and other operations on in-GPU memory matrices. The basic data structure of libcudf is the GPU DataFrame (GDF), which in turn is modeled on Apache Arrow’s columnar data store.
 RAPIDS
RAPIDS
cuDF technology stack
The RAPIDS Python library presents the user with a higher level interface resembling dataframes, like those in Pandas. In many cases, Pandas code runs unchanged on cuDF. Where this is not the case, usually only minor changes are required.
User defined functions in cuDF
Once you’re past basic data manipulation, it is sometimes necessary to process rows and columns with user defined functions (UDFs). cuDF provides a PyData style API to write code to process more course-grained data structures like arrays, series, and moving windows. Currently only numeric and Boolean types are supported. UDFs are compiled using the Numba JIT compiler, which uses a subset of LLVM to compile numeric functions to CUDA machine code. This results in substantially faster run times on the GPU.
Strings in cuDF
Although GPUs are fantastic for rapidly processing float vectors, they haven’t typically been used for processing string data, and the reality is that most data comes to us in the form of strings. cuStrings is a GPU string manipulation library for splitting, applying regexes, concatenating, replacing tokens, etc. in arrays of strings. Like other functions of cuDF, it is implemented as a C/C++ library (libnvStrings) and wrapped by a Python layer designed to mimic Pandas. Although the string data type is not optimized for execution on GPUs, parallel execution of the code should provide a speedup over CPU-based string manipulation.
Getting data in or out of cuDF
Dataframe I/O is handled by a dedicated library, cuIO. All of the most commonly encountered formats are supported, including Arrow, ORC, Parquet, HDF5, and CSV. If you are lucky enough to be running on DGX-2 hardware, you can use GPU Direct Storage integration to move data directly from high-speed storage to the GPU without involving the CPU. Mortal users will still appreciate the speedup the GPU gives when decompressing large data sets, and the tight integration with the Python ecosystem.
GPU Direct Storage is currently in alpha, and when released will be available on most Tesla GPUs. You can create a GPU dataframe from NumPy arrays, Pandas DataFrames, and PyArrow tables with just a single line of code. Other projects can exchange data via the __cuda_array_interface__ for libraries that fall within the Numba ecosystem. DLPack for neural network libraries is also a supported interface.
Probably the biggest drawback in using cuDF is the lack of interoperability outside of Python. I think a focus on a strong foundation of C/C++ APIs, as Arrow has done, would enable a broader ecosystem and benefit the project as a whole.
RAPIDS’ cuML
cuML’s stated goals are to be “Python’s Scikit-learn powered by GPUs.” In theory this means you should only have to change your import statement and perhaps tune a few of the parameters to account for the differences in running on a CPU, where sometimes a brute force approach is better. The benefit of having a GPU-based Scikit-learn is hard to understate. The speedups are substantial, and data analysts can be many times more productive. The C++ API isn’t quite ready for broad consumption outside of its Python bindings, but this is expected to improve.
cuML also includes APIs for helping with hyperparameter tuning via Dask, a library for scaling Python across multiple nodes. Many machine learning algorithms can be effectively made parallel, and cuML is actively developing both multi-GPU and multi-node, multi-GPU algorithms.
 RAPIDS
RAPIDS
cuML algorithms
RAPIDS’ cuGraph
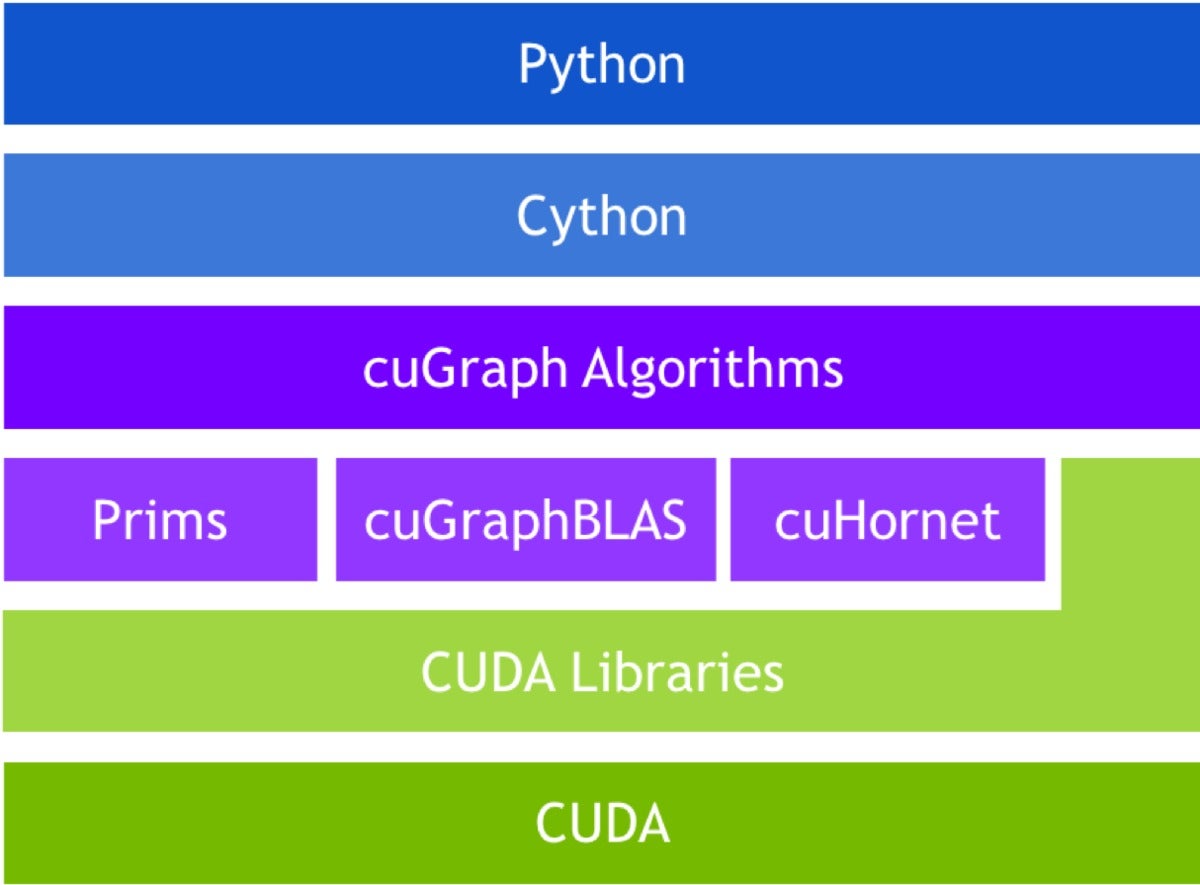
cuGraph is the third member of the RAPIDS ecosystem, and like the others, cuGraph is fully integrated with cuDF and cuML. It offers a good selection of graph algorithms, primitives, and utilities, all with GPU-accelerated performance. The selection of APIs in cuGraph is somewhat more extensive than in other parts of RAPIDS, with NetworkX, Pregel, GraphBLAS, and GQL (Graph Query Language) all available.
 RAPIDS
RAPIDS
cuGraph stack
cuGraph is more like a toolkit in spirit than cuML. Graph technology is a fast-moving space both in academia and industry. Thus, by design, cuGraph gives developers access to the C++ layer and graph primitives, encouraging third parties to develop products using cuGraph. Several universities have contributed, and projects from Texas A&M (GraphBLAS), Georgia Tech (Hornet), and UC Davis (Gunrock) have been “productized” and included under the cuGraph umbrella. Each project provides a different set of capabilities, all GPU-accelerated, and all backed by the same cuDF dataframe.
NetworkX is the Python API targeted by the RAPIDS team for its native interface. There are a number of algorithms available via that interface. While only page rank is multi-GPU, the team is actively working on multi-GPU versions of the others, where applicable.
 RAPIDS
RAPIDS
cuGraph algorithms
One of the cuGraph sub-projects I found interesting is cugraphBLAS, an effort to standardize building blocks for graph algorithms in the language of linear algebra. Based on GraphBLAS (graphblas.org), a custom data structure designed for sparse dynamic graphs processing.
Another cuGraph sub-project, Hornet provides a system independent format for containing graph data, analogous to the way Apache arrow provides a system independent way to process dataframes. Hornet supports most of the popular graph formats including SNAP, mtx, metis, and edges.
In keeping with the spirit of being close to the Python community, Python’s native NetworkX package can be used for the study of complex networks. This includes data structures for graphs and multi-graphs, reimplemented using CUDA primitives, allowing you to reuse many of the standard graph algorithms and perform network structure and analysis measures. The majority of algorithms are single-GPU, like NetworkX. Nevertheless, running them on the GPU alone offers significant speedup, while work continues to move to multi-GPU implementations.
On the RAPIDS roadmap
Given the tremendous speed up that GPU-based analytics provides, there are a few new projects coming into the mix in future versions.
DLPack and array_interface for deep learning
Multi-layer neural networks were one of the first workloads moved to GPUs, and a sizable body of code exists for this machine learning use case. Previously DLPack was the de-facto standard for data interchange among deep learning libraries. Nowadays the array_interface is commonly supported. RAPIDS supports both.
cuSignal
Like most other projects at RAPIDS, cuSignal is a GPU-accelerated version of an existing Python library, in this case the SciPy Signal library. The original SciPy Signal library is based on NumPy, which is replaced with its GPU-accelerated equivalent, CuPy in cuSignal. This is a good example of the RAPIDS design philosophy at work. With the exception of a few custom CUDA kernels, the port to the GPU mostly involves replacing the import statement and tweaking a few function parameters.
Bringing signal processing into the RAPIDS fold is a smart move. Signal processing is everywhere and has many immediately useful commercial applications in industry and defense.
cuSpatial
Spatial and spatiotemporal operations are great candidates for GPU acceleration, and they solve many real-world problems we face in everyday life, such as analyzing traffic patterns, soil health/quality, and flood risk. Much of the data collected by mobile devices, including drones, has a geospatial component, and spatial analysis is at the heart of the Smart City.
Architected like the other components, cuSpatial is a C++ library built on CUDA primitives and the Thrust vector processing library, using cuDF for data interchange. Consumers of the C++ library can read point, polyline, and polygon data using a C++ reader. Python users are better off using existing Python packages like Shapely or Fiona to fill a NumPy array, then using the cuSpatial Python API or converting to cuDF dataframes.
cuxfilter for data visualization
Visualizing data is fundamental, both within the analytics workflow and for presenting or reporting results. Yet for all the magic that GPUs can work on the data itself, getting that data out to a browser is not a trivial task. cuxfilter, inspired by the Crossfilter JavaScript library, aims to bridge that gap by providing a stack to enable third-party visualization libraries to display data in cuDF dataframes.
There have been a few iterations of cuxfilter as the team sorts out the best architecture and connector patterns. The latest iteration leverages Jupyter notebooks, Bokeh server, and PyViz panels, while integration experiments include projects from Uber, Falcon, and PyDeck. This component isn’t quite yet ready for prime time, but is slated for release in RAPIDS 0.13. There are a lot of moving parts, and I did not get to experiment with it first hand, but if it lives up to its promise this will be a great addition to the RAPIDS toolkit.
Scaling up and out with Dask
Dask is distributed task scheduler for Python, playing a similar role for Python that Apache Spark plays for Scala. Dask-cuDF is a library that provides partitioned, GPU-backed dataframes. Dask-cuDF works well when you plan to use cuML or when you are loading a data set that is larger than GPU memory or is spread across multiple files.
Like a Spark RDD (Resilient Distributed Dataset), the Dask-cuDF distributed dataframe mostly behaves just like a local one, so you can experiment with your local machine and move to a distributed model when you need to scale up. Dask-cuML gives cuML multi-node capabilities, making it a good option when you do not have the budget for a DGX workstation.
The Dask project is independent of RAPIDS, and includes several features that a practicing data scientist will find useful, such as low-latency, high-throughput messaging via OpenUCX (normally found in High Performance Computing applications). Dask was made for accessing high-speed block data stores through InfiniBand or large data sets in cloud storage. Dask integration really extends the reach of RAPIDS.
RAPIDS at a glance
RAPIDS addresses one of the biggest challenges of machine learning with Python — slow execution — and does so in an elegant way, by making existing code run on the GPU nearly unchanged. It’s hard to fault the vision or enthusiasm of Nvidia or the community.
While this project has a great deal of momentum, I can’t recommend it for enterprise work at the moment. There are simply too many changes going on under the hood as it evolves. My recommendation for enterprises is “watch this space,” wait for things to settle down, and, if possible, get a few developers involved in contributing. Especially needed are work on making the underlying C++ libraries, especially cuDF, useful for other languages, like R, Java, and Go.
—
Cost: Free open source licensed under Apache 2.0.
Platform: Requires Nvidia Pascal or better GPU with compute capability 6.0 or better and recent CUDA and Nvidia drivers. Supports Ubuntu 16.04, Ubuntu 18.04, CentOS 7, and Docker CE 19.03 or better with nvidia-container-toolkit.
Copyright © 2020 IDG Communications, Inc.






